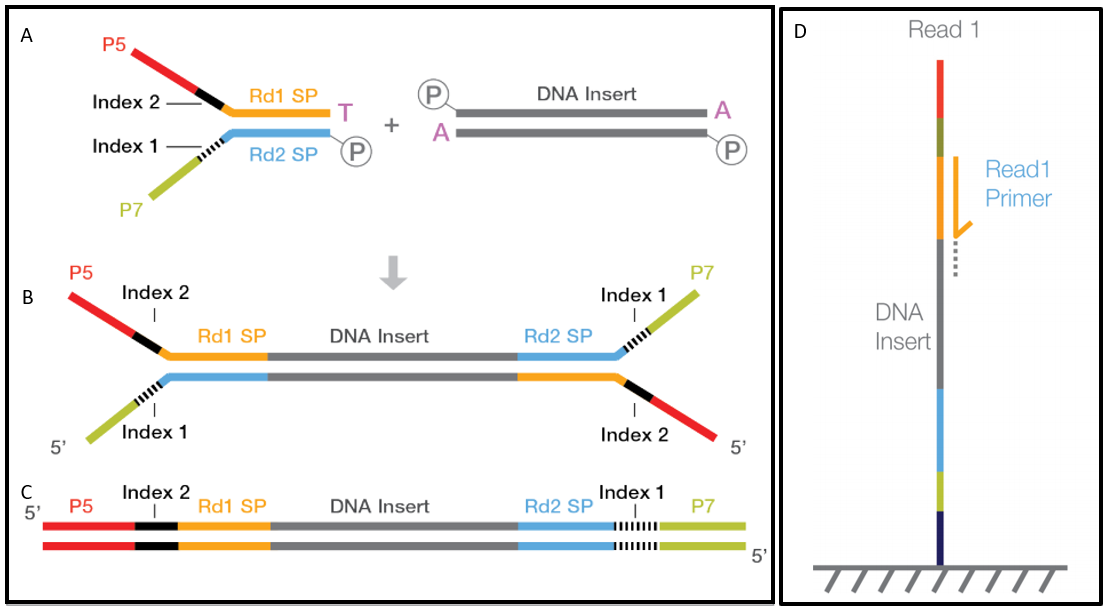
paired end sequencing read length
On sequencing using unpaired reads shows that ultra-short reads theoretically allow whole genome re-sequencing and de novo assembly of only small. Enter up to 20 characters or use manual mode if you need between 20 and 100 bp.
Bulk Rna Seq Data Standards And Processing Pipeline Encode
We use an Illumina MiniSeq for our short-read sequencing runs.

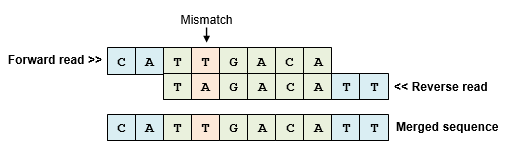
. The pooled indexed libraries were loaded into the MiSeq Reagent kit V2 300 cycles Illumina and sequenced on the MiSeq platform Illumina in paired-end mode with a. The target DNA fragments produced by ART provide the ground truth for the merged paired-end reads. When the fragment length is longer than the combined read length there.
An analysis by Whiteford et al. Modern nextgeneration sequencing platforms offer a range of read configurations such as single-read SR and paired-end PE sequencing with 75 bp per read 100 bp per read and 150 bp. We present the PEAR.
During sequencing it is possible to specify the number of base pairs that are read at a time. We also generated an additional set of 150-bp long reads with a. The library prep protocols are designed to.
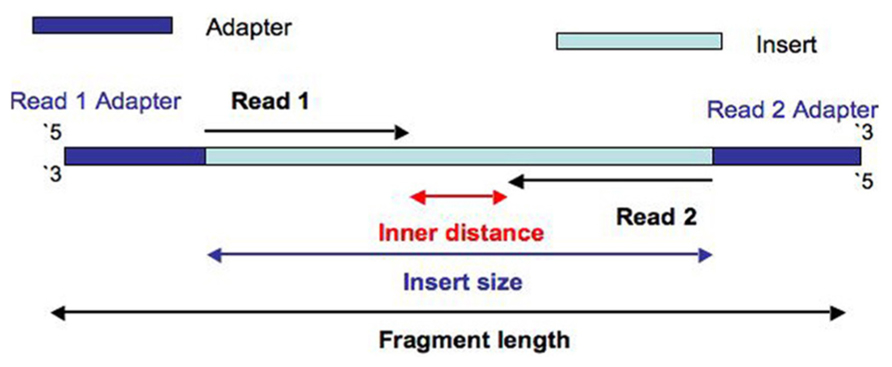
The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phix-musigmaxLp0 where mu is the mean sigma. Simple workflow allows generation of unique ranges of insert sizes. Therefore a robust tool is needed to merge paired-end reads that exhibit varying overlap lengths because of varying target fragment lengths.
Reading the nucleotides from two ends of a DNA fragment is called paired-end tag PET sequencing. However some paired-end sequencing data show the presence of a subpopulation of reads where the second read R2 has lower average qualities. The paired-end short read lengths are always 2 x 150bp 300bp.
Requires the same amount. For example one read might consist of 50 base pairs 100 base pairs or more. To ensure sequencing quality of the Index Read do not exceed the supported.
Why Do Illumina Reads All Have The Same Length When Sequencing Differently Sized Fragments
Direct Chloroplast Sequencing Comparison Of Sequencing Platforms And Analysis Tools For Whole Chloroplast Barcoding Plos One

Five Approaches To Detect Cnvs From Ngs Short Reads A Paired End Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer
How Sequencing Works Ngs Analysis
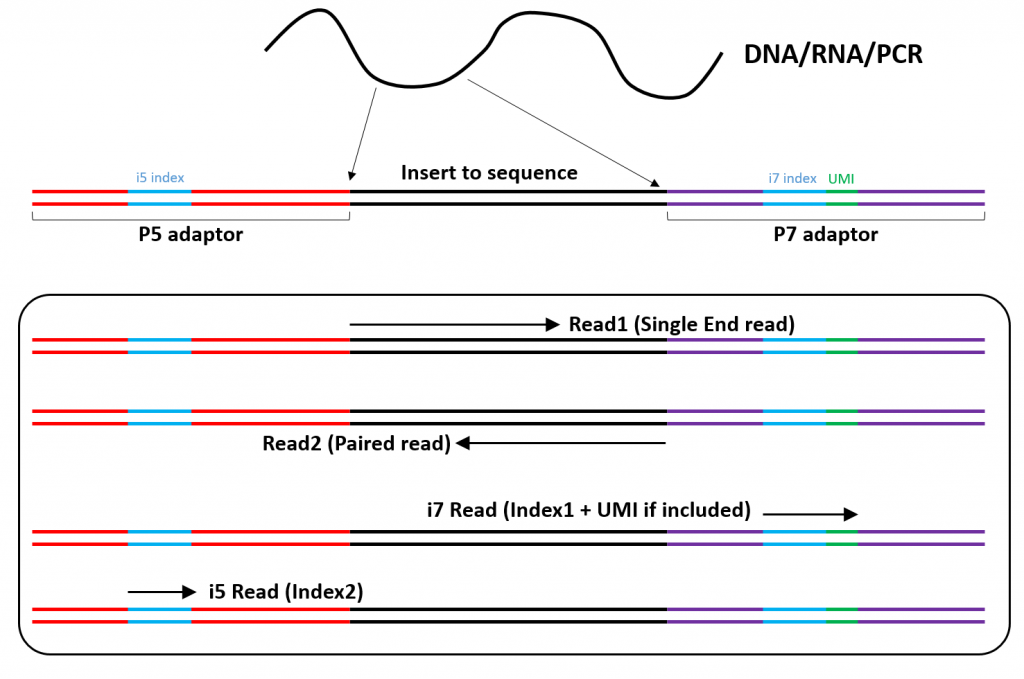
Illumina Short Read Sequencing Lausanne Genomic Technologies Facility
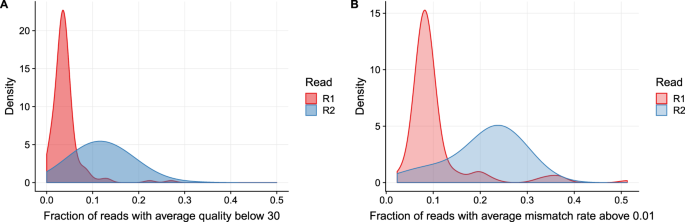
Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Scientific Reports
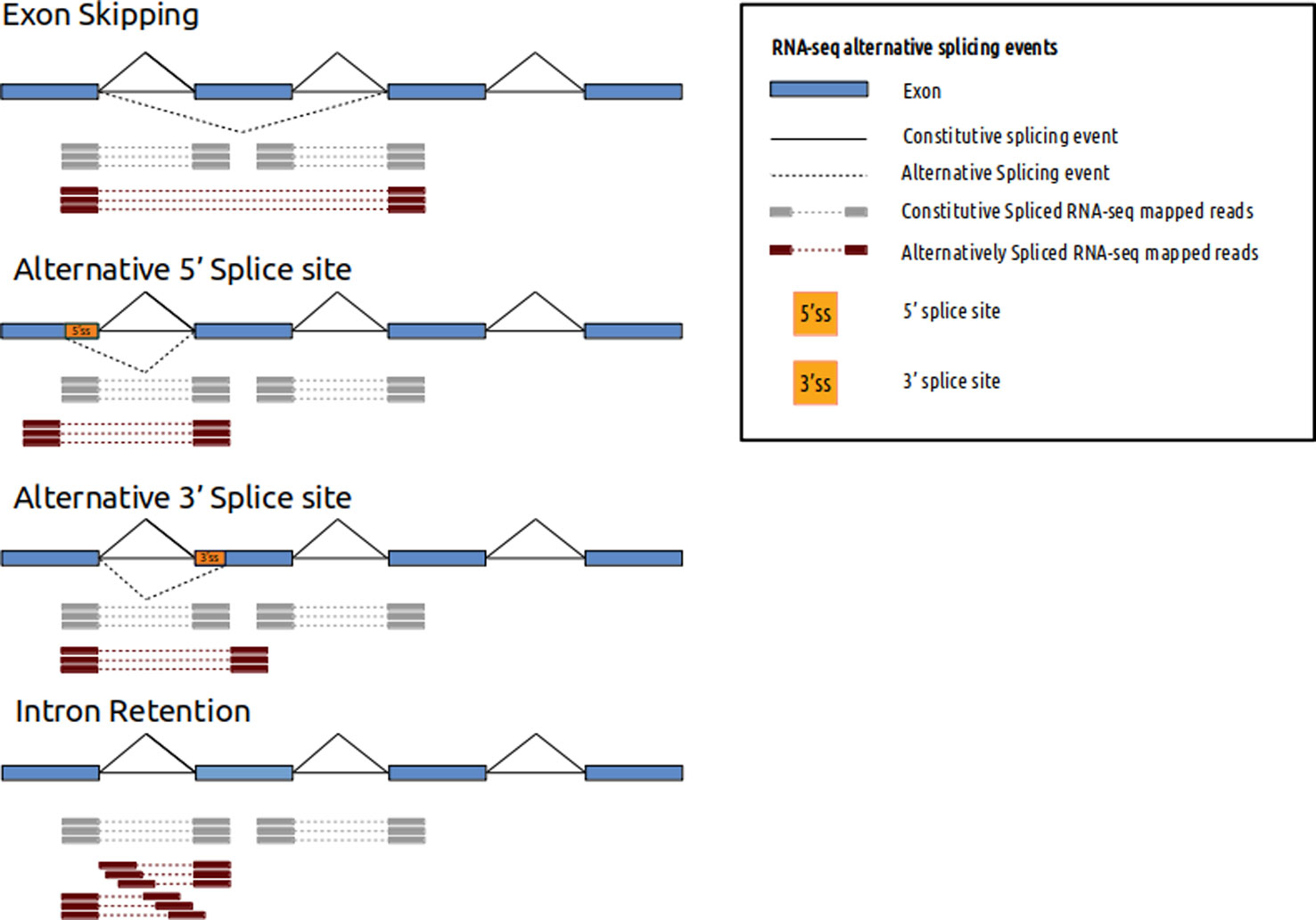
Single End Sequencing Versus Paired End

The Variables For Ngs Experiments Coverage Read Length Multiplexing

Choosing The Right Read Length For Diagnostic Sequencing Cegat Gmbh

How To Calculate The Coverage For A Ngs Experiment

Sequencing Data Analysis Introduction To Key Concepts Youtube

What Are Paired End Reads The Sequencing Center

Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Biorxiv
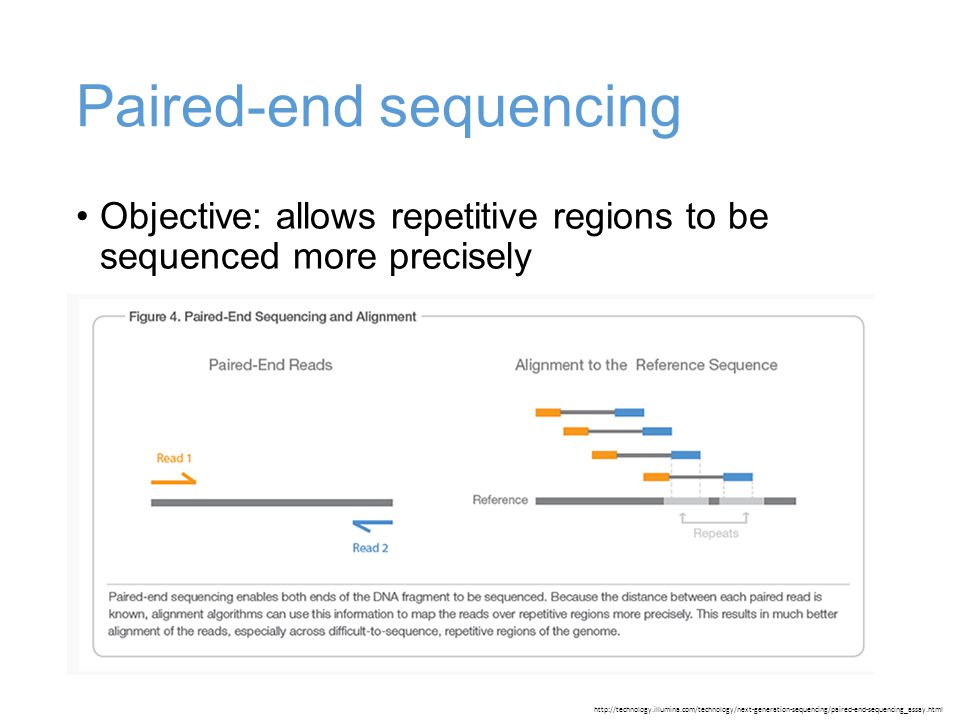
Mcb Lecture 9 Sept 23 14 Illumina Library Preparation De Novo Genome Assembly Ppt Download

Dnbseq G50 Gets Full Performance Upgrade Mgi Leading Life Science Innovation